The 'Archetypes' Project
Q: How many Stable States are there for the Immune System? Over the past few decades, immunologists have characterized the cell types and activation/differentiation status of many immune cells. This has led to important paradigms, such as the ability for T cells to adopt a few fundamental effector states--T helper type 1 (Th1) versus T helper type 2 (Th2) for example. In a given immune site, however, multiple differentiation states typically co-exist, often with one being the more dominant. Presumably the dominant state exists because a collectionof cells 'agree' on this state--through paired gene expression and communication. We call that paired situation (of cells, of gene expression) an archetype. This project interacts strongly with the UCSF ImmunoX CoProjects initiative.
Current: Discovery of Immune Archetypes and the Patterns that Link Them
We aim to define these linked states, both in health and disease. Seeing these will help us manipulate immunity, for example to find the means to inter-convert one kind of immunity for another. In this way, the 'defective' state in one chronic disease may represent the cure in another.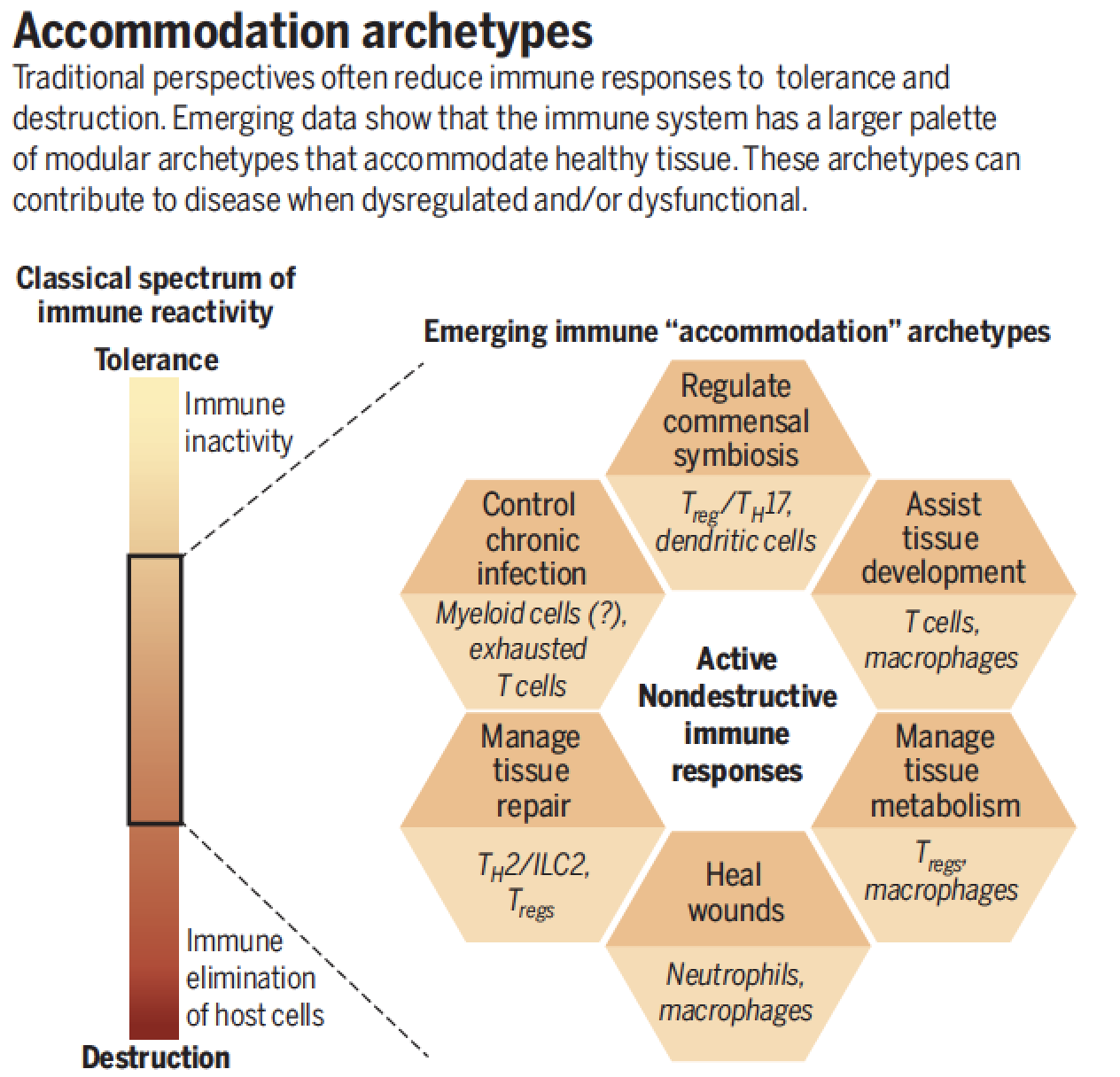
Historical Leadup to this Project
Multicellular Dynamics and ‘Quorum’ decision making in Tissues
Drawing on our experiences with motility and synapses, we have hypothesized that T cells are highly influenced by other cues in their environment and can transmit information amongst themselves via a variety of immunological synapses. This has led to two lines of inquiry into points of information-integration during the course of the immune response.
In the first case, we began to study the effects of migratory cues (i.e. chemokines) upon the ability of T cells to form an immunological synapse and signal. In Friedman et al. Nature Immunology. 2006, by studying the dynamics, we demonstrated that these two systems work synergistically—the chemokine signal helping to hold the cell in place and get it primed to receive the TCR stimulus. We also investigated the role of chemokine signals in generating the ‘stop’ signal described in Jacobelli et al 2004. The mechanism whereby the signals here are integrated at the biochemical level now represents the next step in understanding T cell dynamics in the context of combinatorial signals.
As part of observations of T cell activation in live lymph nodes, we revisited an age-old observation that T cells frequently interact as ‘clusters’. In Sabatos, Doh et al. 2008, we showed that T cells communicate directly with one-another using a synapse-structure, which we term a T-T synapse (see also Doh, and Krummel Current Topics in Microbiology and Immunology, 2010). This synapse increases the specificity of cytokine delivery (such as IL-2) between adjacent activating cells, resulting in increase signaling via the STAT-5 signal-transduction module. In Gerard et al Nature Immunology 2013, we demonstrated that T-T interactions are important for protective CD8 T cell differentiation.
The Allergic Lung
The Immune system is always engaged in surveilling the lung. This involves ongoing ‘sampling’ of the airway contents, as we revealed in Thornton et al. JEM 2012, by a subset of phagocytic cells called dendritic cells. These cells compete with another set of cells, called alveolar macrophages, which also can ingest materials that are breathed in. In Allergic Airway diseases (of which Asthma is an example), direct imaging reveals a bottleneck that forms adjacent to the airway; a meeting site is formed where inhaled particle appears to be ‘presented’ to instigating T cells. This site will be a focus for our ongoing work, in an effort to understand how immunity and tolerance are achieved at this mucosal surface. As part of studying lung immunology, we sought to do so in a fully intact and together with long-time collaborator Mark Looney, devised a method for live-imaging fully ventilated lungs in mice (Looney et al Nature Methods 2011). This method has allowed us to study lung cancer metastasis (Headley et al. Nature 2016) and Marks’s lab to reveal the lung as a site for platelet formation (Lefrancais et al. Nature 2017)
Tumors
In a similar vein, we have approached the direct assessment of immune function in tumor tissues. There is strong reason to believe that the immune dynamics in spontaneous tumors differs considerable from ‘ectopic’ models, the latter being most akin to immunization. To facilitate the study of this, we have undertaken direct imaging of T cells and antigen presenting cells in a mouse model of breast cancer (Egeblad et al. Dis. Model. Mech 2008). This method provides evidence of active surveillance by infiltrating monocyte-lineage cell types as well as suggestive evidence for the inactivation of lymphocytes, which has become an active area of investigation in the lab. More recent publication have served to identify key myeloid players and dynamics that regulate T cell responses (Cancer Cell 2012,2014,2016 and Nature 2016). More details on this approach are in the next theme.
Regulatory T cell Dynamics
In the process of taking the hard-approach (setting up to image T cell receptors, see first theme) we made progress in visualizing T cells within lymph nodes. At the time, a major issue was the dynamics of how regulatory T cell affected T helper synapse dynamics. For that reason, a graduate student in my lab sought to determine how these affected the arrest of T cells in a diabetes model. Together with a postdoctoral fellow in a collaborating lab, they achieved this—showing that regulatory cells inhibit stable interactions of naïve CD4+ T cells with dendritic cells (Tang et al. Nature Immunology. 2006). Another nice aspect of this work was the exploitation of a Mouse-insulin promoter GFP (MIP-GFP) mouse—we crossed this into the genetic background for these studies and were able to use this to identify and image islet-draining dendritic cells using the GFP fluorescence. This formally showed that both regulatory T cells and helper T cells were competing for the same pool of dendritic antigen-presenting cells.
